User Inputs
The user is required to input the protein and ligand between which interactions are to be determined. The user needs to consider the following scenarios to determine which of the two available input options are to be used:
-
User does not want LIFT Visualizer to perform docking on the submitted protein-ligand pair.
Examples of this scenario are when the user wishes to see the interactions in an existing protein-ligand complex. In this case, the user may upload a .PDB file of a protein ligand complex via the “Browse” button (Figure 1, “1”), or enter the PDB ID of a protein-ligand complex from https://www.rcsb.org (Figure 1, “2”).
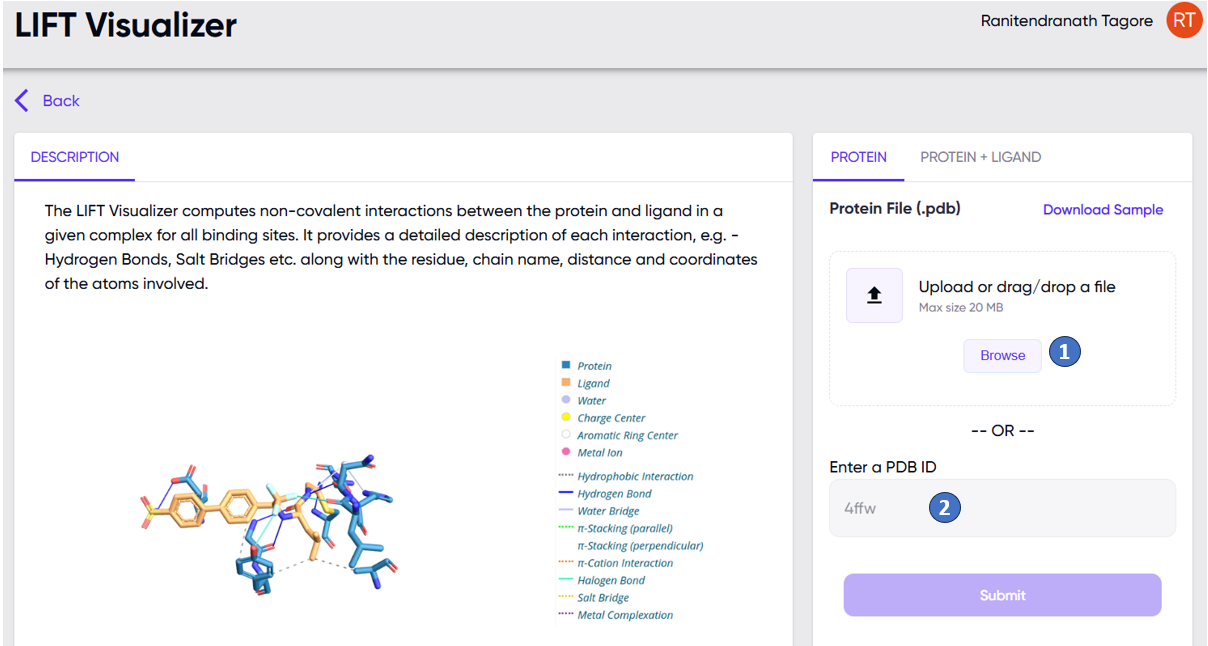
Figure 1. The LIFT visualizer app input screen to be used when docking is not needed (see text). Options to upload a .pdb file (“1”) or the PDB ID from https://www.rcsb.org (“2”) of an existing protein-ligand complex are marked. Note that these options are available under the “PROTEIN” tab of the user input panel.
-
User wants LIFT Visualizer to perform docking on the submitted protein-ligand pair.
In this case, the user needs to specify the binding pocket in which the ligand is to be docked. This is done by uploading, via the “Browse” button (Figure 2, “1”), a .pdb file of the protein of interest, with a ligand occupying the desired binding pocket. The residue name corresponding to the ligand (Figure 2, “2”), and the protein chain to which the ligand is bound (Figure 2, “3”) are also to be specified. Finally, the ligand which is to be docked to the protein is supplied either as a .sdf file (Figure 2, “4”), or as a SMILES string (Figure 2, “5”).
Note that the ligand contained in the .pdf file need not be the same as the ligand supplied later as a .sdf file or a SMILES string. The former is needed only to specify the binding pocket within the protein, while the latter is the ligand on which the actual docking is desired to be performed.
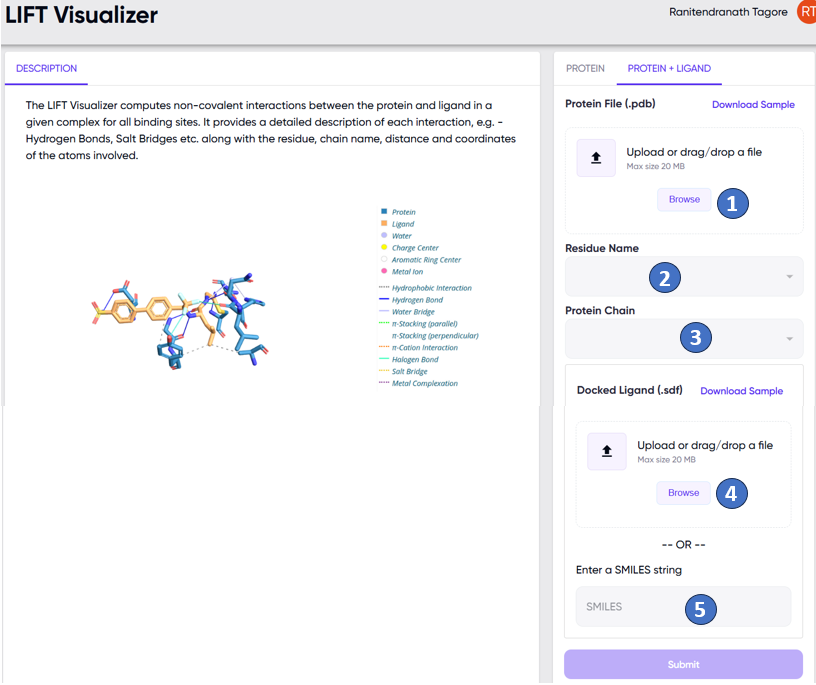
Figure 2. The LIFT visualizer app input screen to be used when docking is desired to be performed by the LIFT visualizer (see text). Options to upload a .pdb file (“1”) of an existing protein-ligand complex, to specify the residue name of the ligand in the complex (“2”), and the protein chain to which the ligand is bound (“3”) are marked. The ligand with which docking is intended to be performed is supplied as a .sdf file (“4”) or as a SMILES string (“5”). Note that these options are available under the “PROTEIN + LIGAND” tab of the user input panel.